
29 Nov Elevate your genomic discoveries with Nanopore Sequencing
What is Nanopore Sequencing?
Nanopore sequencing is a third-generation sequencing technology (TGS) that enables the real-time sequencing of DNA or RNA molecules. It utilizes nanopores, which are tiny protein channels embedded in a membrane, to directly detect and analyze individual DNA or RNA molecules as they pass through the nanopore.
The key principle of nanopore sequencing involves passing a single-stranded DNA or RNA molecule through the nanopore. As the molecule moves through the pore, it causes disruptions in the electrical current flowing across the nanopore. These disruptions are unique to the specific DNA or RNA (modified) bases present in the molecule and are detected by measuring changes in the electrical current.
The process of nanopore sequencing involves the following steps:
- Library Preparation
DNA or RNA molecules are prepared for sequencing by adding specific adaptors or linkers to their ends. These adaptors facilitate the attachment of the molecules to the nanopore sequencing device.
- Sequencing Process
The prepared library is loaded onto the nanopore sequencing device, which consists of a nanopore embedded in a membrane. An electric current is applied across the nanopore, and the DNA or RNA molecules are passed through the pore. As the molecules move through the nanopore, the changes in the electrical current are recorded in real time.
- Base Calling
The electrical current signals are analyzed to determine the sequence of (modified) bases in the DNA or RNA molecule. The disruption patterns in the current are correlated with specific nucleotides (A, C, G, T for DNA; A, C, G, U for RNA), allowing the sequence to be reconstructed and the modifications to be determined ((hydroxy)methylation, acetylation, …).
- Data Analysis
The raw data obtained from nanopore sequencing is processed and analyzed using bioinformatics tools to generate the final sequence data. This includes error correction, alignment to a reference genome or transcriptome, and variant detection, if applicable.
Nanopore sequencing offers several advantages compared to other sequencing technologies. It enables long-read sequencing, allowing the detection of large-scale genomic structural variations, such as insertions, deletions, and repetitive regions, which can be challenging for short-read sequencing technologies. Additionally, nanopore sequencing is a portable and flexible platform, with handheld devices available that provide real-time sequencing capabilities in various settings, including field research and point-of-care diagnostics.
Currently, the error rates are comparable to next-generation sequencing methods (Illumina) and other third-generation sequencing methods (PacBio) obtaining Q20+ or higher to Q30. Ongoing advancements and improvements in nanopore sequencing technologies are still continuously reducing these error rates.
Overall, nanopore sequencing is a powerful and versatile sequencing technology that offers real-time, long-read sequencing capabilities, making it valuable for a wide range of applications in genomics research, clinical diagnostics, and beyond.
What different instruments are available for Nanopore Sequencing?
Output specifications:
| Flongle | MinION | GridION | PromethION 2 | PromethION 24 | |
| Channels/flow cell | 126 | 512 | 512 | 2675 | 2675 |
| Flow cells/device | 1 | 1 | 5 | 2 | 24 |
| Yield (typical) | 1Gb | 15Gb | 75Gb | 300Gb | 3.6Tb |
| Barcoding | 96 | 96/384 | 96/384 | 96/384 | 96/384 |
Note that the generated output is dependent on the sequencing protocol used.
More information can be found on the Oxford Nanopore Technologies website.
What different sequencing protocols are available for Nanopore Sequencing?
DNA sequencing techniques
Nanopore technology can be applied across all DNA sequencing techniques.
- Whole genome sequencing
Accurately characterize all genomic variants and generate complete genome assemblies. - Targeted DNA sequencing
Sequence long targeted regions and expand on the limitations of traditional targeted sequencing approaches. Use amplification-free strategies to preserve base modifications. Or provide a whitelist containing your regions to be enriched with adaptive sampling. - Metagenomic DNA sequencing
More accurately identify microbes and their abundance with real-time results. - Epigenetics
Identify base modifications alongside DNA sequence using direct sequencing approaches resulting in a bimodal readout.
RNA sequencing techniques (transcriptomics):
Nanopore technology enables complete, isoform-level transcriptome characterization.
Using nanopore sequencing, you can:
- Characterise and quantify full-length transcripts
- Unambiguously identify splice variants and fusion transcripts
- Reveal gene expression heterogeneity with single-cell transcriptomics
- Explore epigenetic modifications through direct RNA sequencing
- Enhance viral identification from metagenomic samples (RNA viruses)
For what different applications can Nanopore Sequencing be used?
Nanopore sequencing has found a wide range of applications across various fields of biology and genomics. This transformative technology offers unprecedented insights into the molecular world, enabling researchers to unravel the complexities of genetic and molecular processes.
- Genome Sequencing
Nanopore sequencing is used for whole-genome sequencing of various organisms, including bacteria, viruses, fungi, plants, and animals. Its long-read capabilities help resolve complex genomic regions, repetitive sequences, and structural variations, making it valuable for de novo genome assembly and structural variant detection. - Transcriptomics
As previously discussed, nanopore sequencing is a powerful tool for transcriptomics. It allows researchers to characterize full-length transcripts, identify splice variants, and quantify gene expression accurately. This is crucial for understanding gene regulation and alternative splicing events. - Epigenetics
Nanopore sequencing can directly detect DNA and RNA modifications, including DNA methylation and RNA modifications like m6A. This has applications in studying epigenetic regulation, the effects of modifications on gene expression, and epigenetic variations in diseases. - Metagenomics
Nanopore sequencing is well-suited for metagenomic studies, enabling the identification and characterization of diverse microbial communities in environmental samples, microbiomes, and clinical specimens. It aids in studying microbial diversity, functional potential, and interactions within ecosystems. - Viral Genomics
In virology, nanopore sequencing is used for rapid and accurate viral genome characterization. It’s particularly valuable during viral outbreaks, allowing for real-time monitoring of viral evolution, detection of variants, and assessment of potential drug resistance. - Structural Biology
Nanopore sequencing can be applied in structural biology to investigate the three-dimensional structure of biomolecules. By threading molecules through nanopores, researchers can study the folding and conformational changes of proteins, RNA, and DNA at the single-molecule level. - Clinical Diagnostics
Nanopore sequencing holds promise in clinical diagnostics, especially for identifying genetic mutations responsible for rare diseases and cancers. Its ability to detect structural variants and characterize the entire genome aids in identifying disease-causing variants and potential therapeutic targets. - Environmental Studies
Environmental scientists use nanopore sequencing to study biodiversity in various ecosystems, monitor pollution, and assess the impact of environmental changes on microbial communities. It helps in understanding how ecosystems respond to disturbances and climate change. - Forensic Genomics
Nanopore sequencing can be employed in forensic genomics to analyze DNA samples for identification purposes, especially when dealing with degraded or limited DNA samples. - Evolutionary Biology
Researchers use nanopore sequencing to study evolutionary processes, track genetic adaptations, and investigate the genomic changes that occur in populations over time. - Agriculture and Crop Improvement
Nanopore sequencing is applied in agriculture to study crop genomes, identify disease-resistance genes, and improve crop breeding strategies. - Education and Outreach
Nanopore sequencing has also found use in educational settings, allowing students and researchers to gain hands-on experience with sequencing technology and genomics.
Nanopore sequencing’s versatility and portability have expanded its reach across numerous scientific disciplines, making it a transformative technology for advancing our understanding of biology, genomics, and related fields. Its real-time data generation capabilities make it especially valuable for applications requiring rapid insights and on-site sequencing. Whether it’s deciphering genomes, unraveling the transcriptome, exploring epigenetic modifications, or delving into the mysteries of the microbial world, nanopore sequencing is a powerful tool driving innovation and discovery in the life sciences.
What are the advantages of Nanopore Sequencing?
Nanopore sequencing offers several advantages that make it a valuable tool in genomics research and clinical applications. Here are some key advantages:
- Real-Time Sequencing
Nanopore sequencing provides real-time data generation, meaning that the sequencing process can be monitored and analyzed as it happens. This enables rapid insights into the data, allowing researchers to make decisions on the fly and potentially accelerate the research process.
- Long-Read Sequencing
One major advantage of nanopore sequencing is its ability to generate long sequencing reads. Unlike other sequencing technologies that produce shorter reads, nanopore sequencing can generate reads that span several kilobases in length. This makes it particularly useful for studying large-scale genomic structural variations, such as insertions, deletions, and repetitive regions, which can be challenging to detect with short-read sequencing technologies.
- Portability and Field Applications
Nanopore sequencing devices are available in portable formats, allowing for sequencing to be conducted in various settings, including field research, remote locations, or point-of-care diagnostics. This portability is advantageous for applications such as pathogen detection, environmental monitoring, or rapid response situations where immediate sequencing results are critical.
- Direct DNA/RNA Detection
Nanopore sequencing allows for direct detection and analysis of DNA and RNA molecules without the need for amplification or PCR. This direct detection capability can be advantageous for studying native DNA or RNA molecules, including modified bases, epigenetic modifications, or RNA modifications.
- Real-Time Epigenetic Analysis
Nanopore sequencing has the potential to provide real-time analysis of epigenetic modifications. By detecting the changes in the electrical current as DNA or RNA molecules pass through the nanopore, it can potentially identify and characterize epigenetic marks, such as DNA methylation or histone modifications, in a single-molecule manner.
- Cost Efficiency
Nanopore sequencing can offer a cost-effective alternative to other sequencing technologies, especially for certain applications such as long-read sequencing. The scalability of nanopore sequencing devices and the potential for reusability of the flow cells contribute to its cost efficiency.
- Amplification-free enrichment
Nanopore sequencing offers also a way to enrich certain regions of interest (ROIs) without any PCR amplification step involved. Nanopore adaptive sampling on the other hand, also known as adaptive sequencing, is a feature of nanopore sequencing technology that dynamically adjusts the sequencing parameters based on the characteristics of the DNA or RNA molecules being sequenced. This adaptive approach aims to optimize the sequencing process by focusing on specific regions or types of molecules of interest, thereby improving the efficiency and accuracy of data acquisition.
The advantages of nanopore sequencing make it a powerful tool for a wide range of genomics research, clinical diagnostics, and field applications.
What is adaptive sampling?
The adaptive sampling method is a way to enrich sequencing for certain regions of interest (ROIs) without the need for PCR amplification.
Nanopore adaptive sampling, also known as adaptive sequencing, is a feature of nanopore sequencing technology that dynamically adjusts the sequencing parameters based on the characteristics of the DNA or RNA molecules being sequenced. This adaptive approach aims to optimize the sequencing process by focusing on specific regions or types of molecules of interest, thereby improving the efficiency and accuracy of data acquisition. Adaptive sampling can be seen as an in silico enrichment method.
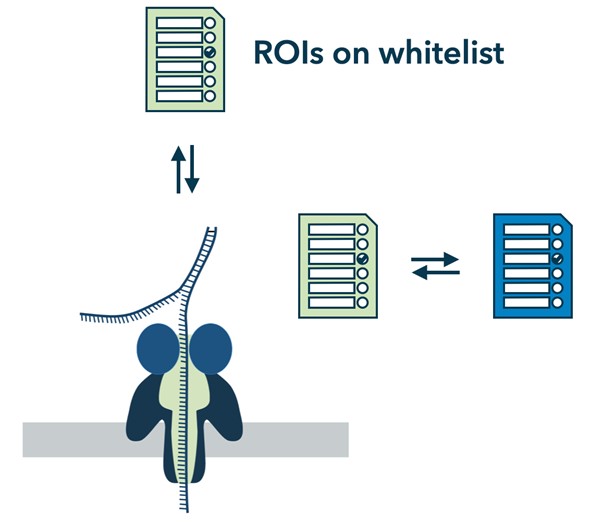
The concept of nanopore adaptive sampling involves two key components:
- Real-Time Analysis
During the sequencing process, as DNA or RNA molecules pass through the nanopore, the changes in the electrical current are recorded in real-time. This real-time data acquisition enables the analysis of the electrical signal as the molecule is being sequenced.
- Signal Analysis and Decision-Making
The electrical signal generated during nanopore sequencing contains information about the sequence and characteristics of the DNA or RNA molecule. By analyzing this signal in real-time, algorithms and software can make decisions about the molecule’s characteristics, such as length, quality, or identity, and adjust the sequencing parameters accordingly.
Based on the real-time analysis and decision-making, nanopore adaptive sampling can be used to achieve different goals, such as:
- Quality Control and Error Correction
The adaptive sampling approach can help identify sequencing errors or poor-quality reads as they occur. By monitoring the signal characteristics, algorithms can dynamically decide whether to continue sequencing a particular molecule or terminate it early if it is likely to be erroneous or of low quality. This helps improve the overall accuracy and quality of the sequencing data.
- Targeted Region Sequencing
Adaptive sampling can be used to selectively enrich specific regions of interest within the genome. By analyzing the signal, algorithms can recognize specific features or patterns associated with the target regions. The sequencing parameters can then be adjusted to increase the coverage and accuracy of the reads within those regions, while reducing sequencing efforts in non-targeted regions.
- Epigenetic Modification Detection
Nanopore adaptive sampling has the potential to identify and analyze epigenetic modifications in real time. By monitoring the electrical signal characteristics, algorithms can detect specific patterns associated with DNA or RNA modifications, such as DNA methylation or RNA modifications. This real-time detection provides a unique opportunity for studying epigenetic modifications at a single-molecule level.
Nanopore adaptive sampling is an ongoing area of research and development in nanopore sequencing. It aims to improve the efficiency and effectiveness of the sequencing process by dynamically adapting to the characteristics of the molecules being sequenced. By optimizing data acquisition and focusing on specific regions or types of molecules, adaptive sampling contributes to the overall advancements and capabilities of nanopore sequencing technology.
Is Nanopore Sequencing accurate?
The read accuracy of nanopore sequencing has significantly improved over the years being currently comparable to other sequencing platforms.
Typically, with the baseline Q20+ chemistry, the raw single-read accuracy of nanopore sequencing is >99%. This means that for each individual base in a sequencing read, there is a chance of 1% that an error may occur. These errors can include insertions, deletions, or substitutions of nucleotides.
It’s important to keep in mind that nanopore sequencing is a rapidly evolving technology, and ongoing advancements, research, and development efforts are continuously addressing the challenges of accuracy and improving the performance of nanopore sequencing platforms. Currently, next to the simplex methods, duplex sequencing is also becoming available leading to Q30+, 99.9% accuracy.
Nanopore Sequencing services with OHMX.bio?
At OHMX.bio, we are proud to offer cutting-edge Nanopore sequencing services in the fields of genomics, epigenomics, and transcriptomics, providing a wealth of information that can revolutionize your research.
Through our TGS (Third-Generation Sequencing) technology, we deliver high-throughput routines that enable rapid and accurate sequencing of various samples. Our protocols and pipelines have been meticulously designed to cater to your specific needs, ensuring precise results and comprehensive insights.
In genomics, our Nanopore sequencing services excel in genome assembly, full 16S or metagenome analysis (microbiome), and quality control for different biologicals (plasmids, viral vectors, recombinant cells). For a focused approach, our targeted sequencing services provide adaptive sampling and Cas9 enrichment, empowering you to zoom in on specific regions of interest without the need for amplification.
In transcriptomics, our Nanopore sequencing services excel in identifying and quantifying full-length transcripts, allowing you to delve deep into gene expression and regulation. We go further by facilitating differential gene expression analysis, enabling you to pinpoint critical differences and uncover valuable markers.
With OHMX.bio, you can embark on groundbreaking research projects and tackle some of the most challenging questions in the fields of genomics, epigenomics, and transcriptomics. Our Nanopore sequencing services offer the precision and scale required to push the boundaries of scientific discovery.
Feel free to reach out to our team with any inquiries or to discuss potential collaborations. Together, we can unlock the full potential of Nanopore sequencing and drive scientific advancements that benefit us all
Do you have questions about Nanopore Sequencing?
Fill out the form below and our experts will get back to you as soon as possible!



