
07 Feb White paper: quality control of biologics – precision safety through long-read sequencing
Abstract
Biologics are nowadays often the result of gene editing, transfections, or other advanced biotechnical methods. As these biologics are administered directly to patients, e.g. through advanced cell & gene therapies, quality control (QC) is one of the, if not the, most important aspect in the development process and manufacturing of these biologics. However, the methods used for QC are often dated and cannot be considered precise, focusing more on consequences than causes, using patient monitoring and flow cytometry. Sequencing, and more specifically long-read sequencing, offers a precise solution. By sequencing edited or transfected cells, the actual genetic code can be monitored for variants or indels (genetic stability), copying, frameshifts, or other unwanted changes (structural variations), as already suggested in the most recent ICHQ5A(R2) guidelines. In this paper, we discuss the (dis)advantages of long-read sequencing as a means of quality control for biologics, as well as discuss three case studies where it already proved useful.
Introduction
Biologics, encompassing a diverse array of therapeutic agents derived from living organisms, have emerged as a key component of much of our modern medicine. For example, insulin was one of the first biologics to be produced on a large scale (extracted from pig pancreas). However, if we think of biologics today, we think of plasmids, viral vectors, transfected cell lines, immune cells (CAR T), or even microbiomes.
Per definition, biologics are drugs produced from living organisms or contain components of living organisms.
These can be delivered in the form of vaccines, blood and blood components, allergenics, somatic cells, gene therapy, tissues and recombinant therapeutic proteins. The applications of biologics are diverse, ever-expanding and range from oncology and autoimmune diseases to infectious diseases, organ transplants and much more. Especially with the advent of advanced biotechnological techniques, the landscape of biologics has witnessed a transformative shift.
However, there is also an inherent risk in producing biologics. On the one hand, this is because the production involves living organisms, often cell lines, which are susceptible to contamination from various agents. On the other hand, techniques which involve the modification of the genetic code (through editing or insertion) depend on the integrity of that part, as well as the stability. For example, Cas9 gene editing can yield off-target activity (1) and several vectors, using promoters of cellular housekeeping genes, have shown unwanted polyclonal vector distribution (2).
The crux of the matter is that, while during the development phase of a biologic drug the custom vectors, plasmids, nucleic acids, … are checked rigorously; once in production the integrity is rarely checked precisely, but rather focusses on downstream characteristics (e.g. protein structure, fragmentation, etc..). This is quality control after the fact as, likely, the inherent cause of the characteristic can already be found in the (epi)genetic sequence and will be detected there with much higher probability. To effectively reduce the chances of a faulty biologics component appearing in generation 10, one should have checked the genetic code aggregating mistakes in generations 1-9.
The only precise solution is to deploy sequencing in quality control.
Sequencing allows the confirmation of the (epi)genetic code, insert sites and insert copy number. Furthermore, it can be used to assess potential contamination with identification of the contaminant without having to use in vivo models.
Sequencing has become common practice and prices have dropped accordingly. Yet, sequencing is not widely adopted in the quality control of biologics. Potentially this is because often sequencing is done using traditional Next-Gen Sequencing. These short reads pose an important drawback when dealing with biologics, as the inserted or modified parts are too long to fit in one read. Short read sequencers (from Illumina or Element Bioscience) generally produce reads with sizes varying from 50 to 300bp. Overall, these are too short for the questions raised related to genetic identification or characterization of biologics.
Long-read sequencing avoids this issue.
Long-read sequencing, as the name implies, sequences longer strands (on average around 15000 bases). Because of this, flanking regions are always included, allowing for an accurate mapping of the insert onto the reference sequence. Furthermore, the long reads can also reliably identify unique insertions (3) due to the spanning of the insert, thus including flanking regions which allow the insert to be uniquely identified. On top of that, long-read sequencing often starts from easier and faster library preparation protocols and is more portable. On the downside, it needs higher input amounts, which are often also required to be of high molecular weight.
Below, we discuss three case studies – a plasmid, a human genomic insertion and a viral vector, – where long-read sequencing was employed to perform quality control of a biologic component. In all three cases, long-read sequencing had clear advantages and some previously unknown quality issues were discovered.
Case studies
Case study 1: plasmid sequence QC
For a plasmid biologic product, containing recombinant genes or an RNA vaccine template, it is important to have an exact understanding of its sequence. This is even more the case when this biologic product goes into a production phase. Therefore, optimal sequence typing methods are of notable significance. Sanger sequencing is for this application still often preferred over next-generation sequencing due to the longer read lengths (700 – 1 000bp), which benefits assembly of the full biologic product. ONT long read sequencing read lengths (with averages typically around 10-20kB) go another step further and as the latest ONT technology reaches Q20+ accuracy, ONT sequencing seems to become the undoubtful go-to sequencing technique for this application.
To demonstrate this application, DNA of a set of plasmid products (with an estimated length of around 6kB) was barcoded, pooled and sequenced together on a MinION R10.4.1 flow cell for 2 hours. After basecalling and demultiplexing, the overall dataset was analyzed in general data QC. This showed that almost all reads cover the full product with an average read length of around 6kB. Furthermore, extrapolation of the observed coverage showed that, even on a run of only 2 hours, one should be able to pool and sequence 96 samples together, obtaining at least 250x sequencing coverage per sample. This is more than enough to allow successful sequence assembly of almost all products.
The read output is initially used to assemble a gross structure of the plasmid (5). This raw assembly is afterwards annotated (10) to pick up the present elements and plot a general structure of the sequence product (Figure 1).
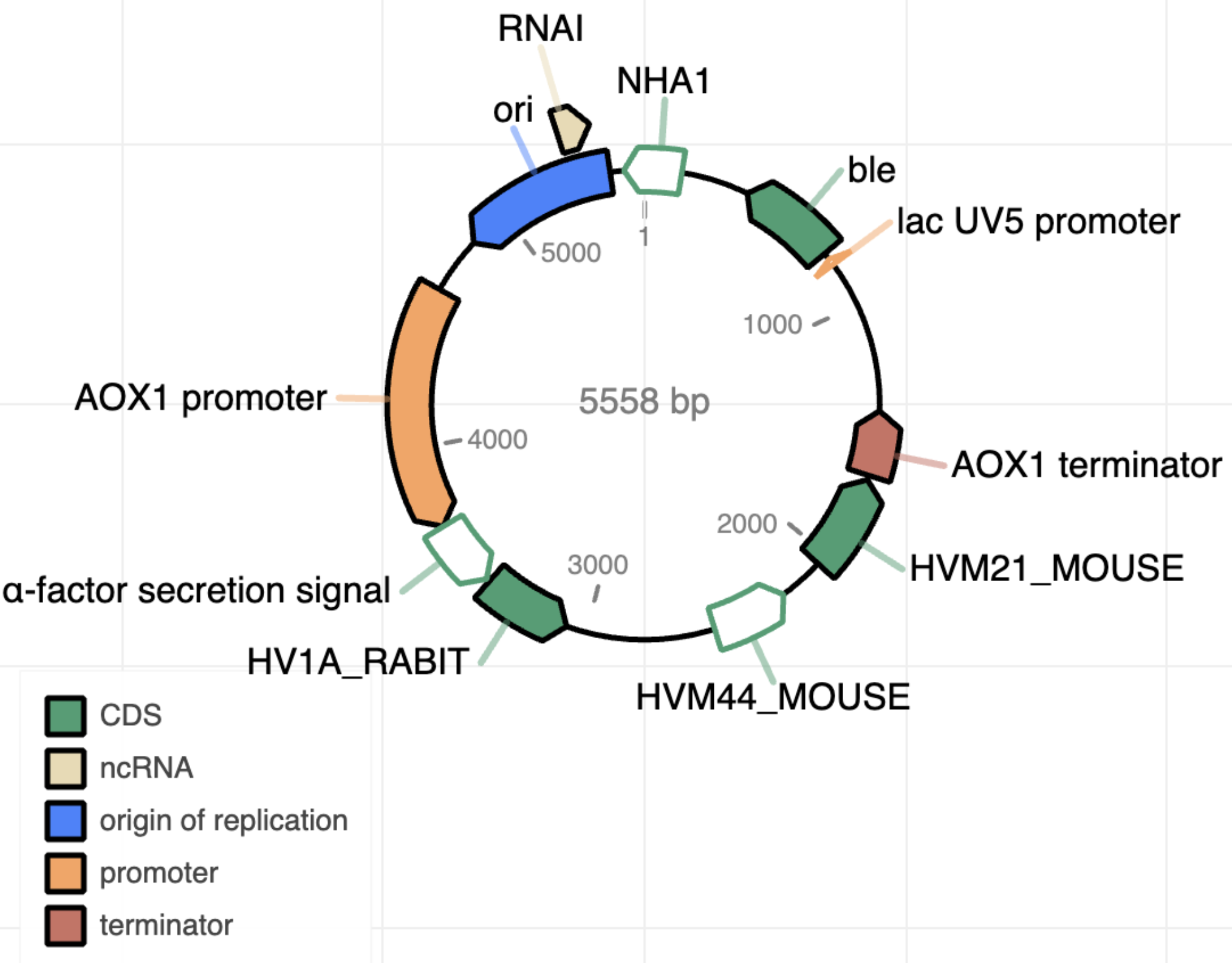
Figure 1 | Example of the output after plasmid annotation.
The assembly strategy could be further improved. Trycycler (9) provides a semi-automatic approach to divide the full read set into subsets, resulting in temporary assemblies per subset. These (sub)assemblies can then be combined into a single final assembly, which is more accurate than the one obtained from all reads at once. Downstream polishing with Medaka can further correct some base-errors in the final assembly result.
On top of the existing Trycycler approach, several custom steps were added specifically for circular plasmids. By checking the different subset assemblies against each other in the light of circularity features, gross structure errors occur less frequently in this custom approach than when a toolset designed for linear products is applied.
All analyzed plasmids contained an introduced array of small custom peptides with polypeptide linker sequences in between the custom peptides. After running the optimized circular plasmid assembly pipeline on the generated ONT data, the found insert was translated into amino acid sequences. For all analyzed products, the sequence matched exactly the expected peptide sequences.
Conclusion: the combination of ONT long read sequencing and a custom bioinformatics pipeline leads to complete and highly accurate plasmid sequence identification.
Case study 2: transfected cell line QC
ONT long read sequencing is also extremely useful in the analysis of larger biologics such as complete modified cell lines. To demonstrate this, a transfected human cell line was analyzed, where a custom reporter block was introduced at unknown locations in the genome. The exact structure of the reporter block was unknown, except for the used promotor sequence and the fact that (the open reading frame of) a reporter gene and several repetitions of a transcriptional response element were present in the reporter block.
Libraries were prepared from DNA of these modified human cells and sequenced on a PromethION R10.4.1 flow cell. After basecalling and QC, it was found that the overall genome coverage was around 52X, sufficient for consensus sequence determination. To get a first rough structure of the inserted reporter sequence, a structural variant calling (8) was performed over the full human genomic sequence. All called variant sequences were afterwards filtered for containing homology (11) with the sequence of the promotor sequence that should be present in the reporter block. Based on these filtered variant sequences, a rough assembly of the reporter block could be constructed (7). The rough assembled sequence of the reporter block was added to the human genome as an artificial additional chromosome. All reads that then mapped to this reporter block sequence (4), were isolated. In this stage, the long read lengths were key for this analysis, as most reads not only cover the full reporter block, but they also contain a lot of information about the neighboring region in the genome where the reporter block was inserted. This neighboring information allowed to determine the genomic insert positions of the reporter block at base-specific level. As such, exactly 13 reporter insertion events (Table 1) were tracked down. This number of insertions could also be confirmed by calculating the ratio between the read coverage over the reporter and the coverage over the overall genome. On the other side, the long reads allowed the assembly of a detailed nucleotide sequence of the reporter block itself (5). With the sequence of the reporter known, also the number of transcriptional response elements, the type of reporter gene and the location of all these elements in the reporter block were determined (Figure 2).
Table 1 | Overview of all found insert positions of the reporter block in the human hg38 genome.
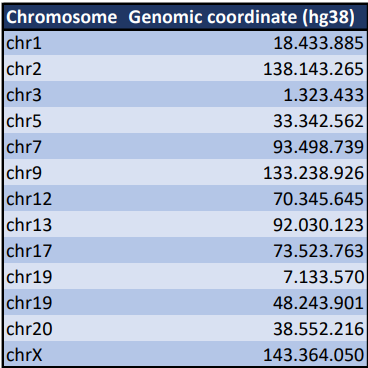
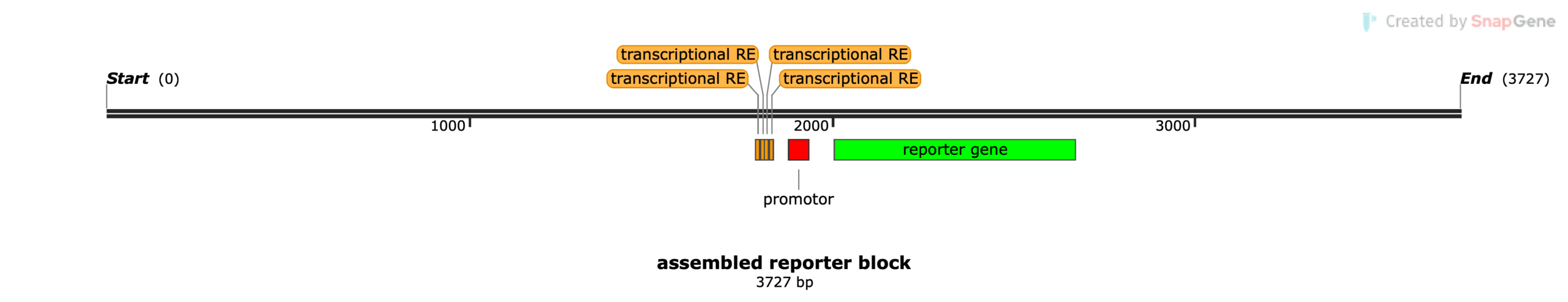
Figure 2 | Found structure for the reporter block.
Conclusion: Given that some sequence characteristics of the inserted reporter are known, these can be used on ONT data to accurately locate integration sites and reveal the full reporter sequence structure using our custom in-house developed pipeline.
Case study 3: sequence stability of a viral product
Nanopore (ONT) long-read sequencing forms an elected tool for investigating the stability of a sequence product between different batches or operations. In this context, a viral product, originally originating from Herpes simplex virus (HSV) genomic DNA and modified with certain inserts and deletions, was analyzed from different batch samples. From a master viral seed stock (MVSS), a working viral seed stock (WVSS) was created, and this stock was at the same time sampled as base level for this stability study. With viral material from this WVSS, four consecutive infection cycles were performed in host cells. From each infection cycle batch, a sample was taken. In summary, five samples were taken and investigated for stability.
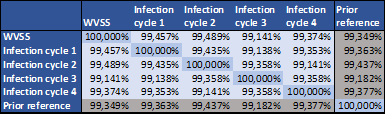
DNA of all samples was isolated, fragmented, quality checked and selected for size. Afterwards, DNA libraries were prepared, barcoded, pooled, loaded and sequenced on MinION R9.4.1 flow cells (the most recent ONT flow cell type at the time of the experiment). The raw sequencing signal was basecalled, demultiplexed, checked for raw data quality and then mapped to a previous reference sequence of the viral product. Thereafter, an assembly was performed per sample, generating a consensus sequence that represents the present viral genome per sample. For this step, one of two approaches can be chosen. The first approach is de novo (5). This approach is suited when large structural variants can be expected. However, de novo approaches often have difficulties in producing one contiguous consensus sequence, especially when there are multiple repetitions or homologue sequences present in the genome. The second approach, reference-based assembly, can produce one contiguous result much easier as it stays close to provided prior reference. In this case, the gross structures of the consensus results were expected to be very similar to the prior reference and therefore, a reference-based assembly was preferred. For this strategy, Racon (6) assembly was combined with Medaka (an open source ONT tool) polishing. Consensus sequences of the different samples and the prior reference were aligned (7) against each other to produce a pairwise identity matrix (Table 2). Differences in identity statistics between the different samples in this matrix are marginal, meaning that these differences will only be effects of sequencing and assembly techniques, rather than genuine sequence differences between samples. As the identity percentages are comparable between infection cycles and with the prior reference, it could be decided that the sequence product is stable over the different batches. These identity percentages could also be constructed for specific regions of interest.
Table 2 | Pairwise identity matrix for all analyzed samples and the prior reference.
Although consistent structural variants were not present in the assemblies, it was interesting in this case to look for more specific small variants and variants present in specific allelic distributions, i.e. heterogenic variations that are present in a subset of the sample’s read material. Alignment statistics per genomic position were piled up, showing an extra nucleotide in a guanine homopolymer stretch in a promotor sequence of interest. Interestingly, this inserted nucleotide was detected in around 30-32% of the reads, showing that this insertion is heterogenically present in all investigated samples in a constant frequency. Similarly, two deletions of around 3 and 7kB were detected in around 13% of the reads for all samples using Sniffles2 (8), a state-of-the-art tool for detecting large structural variants in long-read data. In a later stage, all collected Nanopore long-read data was used to generate a new reference sequence. For the assembly of this new reference, Racon assemblies of the different samples were combined with the Trycycler tool (9). Trycycler allows to construct an even more accurate consensus from different input assemblies of the same genome. Medaka polishing was afterwards applied on this Trycycler assembly. It could be confirmed that the accuracy of this new reference slightly improved over the prior one (Figure 3).
Taken together, while the sequence remained relatively stable over different infection cycles, we did detect some variations present in low allelic frequencies which are currently being further investigated.
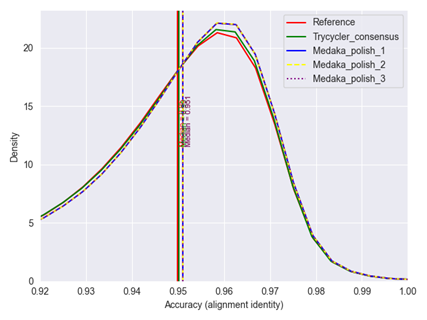
Figure 3 | Accuracy distribution of the different assemblies generated for all sequenced samples. The Trycycler consensus has a slight improvement over the prior reference. Medaka polishing further improved this accuracy. Overall, there was no difference between one or multiple iterative rounds of Medaka polishing. Accuracies are consistent with R9.4.1 flow cells.
Conclusion: ONT long-read data allows for a more accurate determination of stability over different production cycles of a viral product. In the same analysis, low-frequency variations can be detected.
Other advantages of long-read sequencing
Epigenetics
Long-read sequencing has additional features not found in traditional next-gen sequencing. For instance, nanopore sequencing has the ability to simultaneously detect modified nucleotides, such as methylation and hydroxymethylation, while generating long-reads. This is done without the need for e.g. bisulfite treatment, which can highly degrade DNA (12). This multi-omic/bimodal genetic and epigenetic data is generated for every read and enables a deeper understanding of the downstream regulation of the genes, especially when combined with RNA sequencing and/or proteomics.
Adaptive sampling
Adaptive sampling or targeted sequencing is a method unique to nanopore sequencing. In essence, it allows a strand that is being sequenced to be evaluated in real-time. In combination with an inclusion (or exclusion) list, a sequencer can be programmed to only continue sequencing if the first ±400 bases are included in the list (within a margin of error). The main advantage is that, given that certain genes are targeted, reads will mostly be consumed by the targets of interest. This leads to much deeper coverage for those genes for roughly the same cost. The increase in coverage can be up to 14 times compared to a setting without adaptive sampling (13). A clear use for adaptive sampling is for gene complexes, such as the HLA complex or a panel of disease- or cancer-specific genes.
Conclusion
The term precision medicine is often used to describe biologics-based therapies. However, the lack of precise control of the genetic makeup of many biologics is cause for concern. Long-read sequencing is ideally suited to bridge this gap, especially with the recent technological advances delivering the highest quality readouts so far (Q20+). The long readouts guarantee the inclusion of flanking regions, allowing precise localization of indels and variations. Furthermore, specific tools can be deployed to investigate circular plasmids without error-prone assemblies. And apart from the obvious advantages, long-read sequencing can also help explore certain epigenetic mechanisms, and with the inclusion of adaptive sampling, it’s possible to focus resources on only the regions of interest, thereby delivering qualitative results for a smaller price tag.
References
- Pacesa, M., Lin, C. H., Cléry, A., Saha, A., Arantes, P. R., Bargsten, K., Irby, M. J., Allain, F. H. T., Palermo, G., Cameron, P., Donohoue, P. D., and Jinek, M. (2022) Structural basis for Cas9 off-target activity. Cell 185, 4067-4081.e21
- Cavazzana, M., Bushman, F. D., Miccio, A., André-Schmutz, I., and Six, E. (2019) Gene therapy targeting haematopoietic stem cells for inherited diseases: progress and challenges. Nature Reviews Drug Discovery 2019 18:6 18, 447–462
- Pollard, M. O., Gurdasani, D., Mentzer, A. J., Porter, T., and Sandhu, M. S. (2018) Long reads: their purpose and place. Hum Mol Genet 27, R234–R241
- Li, H. (2018) Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100
- Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019) Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37, 540–546
- Vaser, R., Sović, I., Nagarajan, N., and Šikić, M. (2017) Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27, 737–746
- Lassmann, T., and Sonnhammer, E. L. L. (2005) Kalign – An accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6,
- Sedlazeck, F. J., Rescheneder, P., Smolka, M., Fang, H., Nattestad, M., Von Haeseler, A., and Schatz, M. C. (2018) Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods 15, 461–468
- Wick, R. R., Judd, L. M., Cerdeira, L. T., Hawkey, J., Méric, G., Vezina, B., Wyres, K. L., and Holt, K. E. (2021) Trycycler: consensus long-read assemblies for bacterial genomes. Genome Biol 22,
- McGuffie, M. J., and Barrick, J. E. (2021) PLannotate: Engineered plasmid annotation. Nucleic Acids Res 49, W516–W522
- Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36,
- Mill, J., and Petronis, A. (2009) Profiling DNA methylation from small amounts of genomic DNA starting material: efficient sodium bisulfite conversion and subsequent whole-genome amplification. Methods Mol Biol 507, 371–381
- Martin, S., Heavens, D., Lan, Y., Horsfield, S., Clark, M. D., and Leggett, R. M. (2022) Nanopore adaptive sampling: a tool for enrichment of low abundance species in metagenomic samples. Genome Biol 23, 1–27
Do you have questions about long-read sequencing?
Fill out the form below and our experts will get back to you as soon as possible!
