
10 Jul Fishing for the RNA that matters. Adaptive sampling: sequence what is of interest
With its inventive method of sequencing a single nucleoacid strand by passing it gradually through a pore, Nanopore sequencing’s long read technology is revolutionizing the omics world. Adaptive sampling is a clever strategy that allows to enrich or deplete sequencing signal for a selected list of regions of interest in Nanopore sequencing [1]. This is achieved by basecalling and mapping the reads in real-time during the sequencing process with the fastest available models. This process allows to decide whether a nascent read is of interest after collecting signal from the first few hundred bases. If a read is not of interest, the voltage over that specific pore will be reversed, allocating sequencing time of the pore to a more interesting read and as such acquire enrichment on the configured regions of interest (Figure 1). As this strategy only involves programming the sequencing run in the correct way, it makes it a very elegant enrichment solution without any required adaptation in the wet lab preparation.
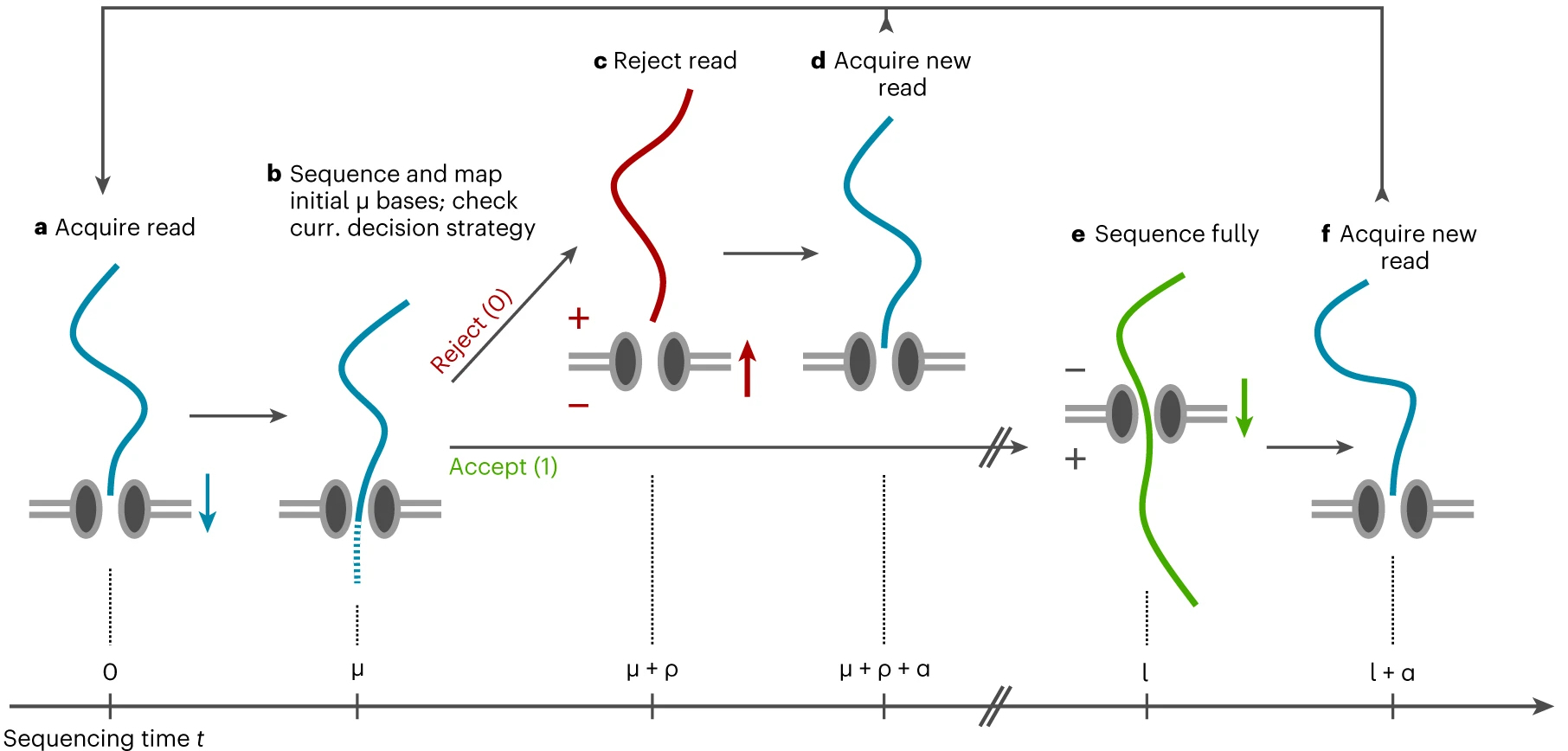
Figure 1: Adaptive sampling enables to obtain enrichment on regions of interest by spending valuable sequencing time rather on fragments of interest. A sequence fragment that is not of interest, will be rejected from the pore after reading the first few hundred bases followed by a decision based on the live mapping results. Figure adapted from Weilguny 2022 [2].
Adaptive sampling is gradually becoming more known for the enriched sequencing of specific regions in DNA. To keep up a descent enrichment factor, an optimal white-listed target set comprises 1-5% of the total (meta)genome of the sample of study. Depending on how much sample material there is available and on how comprehensive the target set is selected, an average enrichment factor of around 4 can be expected for DNA [3].
The interest of adaptive sampling is in what is transcribed
Often, the research interest goes beyond what is present in the DNA. RNA is a much more functional molecule, given its function as information carrier in expression (mRNA) or being active on its own in the form of rRNAs, tRNAs and especially in the form of different other classes of non-coding functional RNA like lncRNA. The RNAs of a specific gene often occur in the form of different co-existent isoforms, depending on how the pre-mature RNA was spliced after transcription. Quantification of the distribution of the different isoforms is often of interest. On top of that, RNA read material can be used to pick up novel unknown isoforms [4].
Similar to what happened in epigenomics a few years ago, RNA research is currently gathering deeper insights in the role of RNA modifications. The field was coined with the term ‘epitranscriptomics’ and currently m6A, m5C and pseudouridine (Ψ) are the modifications getting the most interest. It is becoming more and more clear that these modifications on the RNA affect RNA stability, translation and splicing, impacting gene regulation with applications in development and disease [5].
Nanopore sequencing is causing a revolution for the moment in RNA research by allowing long-read sequencing of native full-length RNA molecules, thereby allowing a direct and more comprehensive analysis of abundance, isoforms, splicing and RNA modifications in one sequencing run. By outknocking reverse transcription, biases and errors are greatly reduced, leading to a new powerful tool for advancing our understanding of gene expression and regulation in medicine and agriculture. On top of that, direct RNA sequencing possesses useful advantages for the metatranscriptomic profiling of microbial communities [3]. The application of adaptive sampling for enriching RNA molecules of interest was withheld for a while though. This was mainly caused by the fact that the theoretical enrichment factor for RNA is several times lower than for DNA. Especially the fact that the translocation speed of RNA through the pore is way slower (70 bases per second for direct RNA compared to 400 for DNA), brought the theoretical enrichment factor down to around 1.4 [6].
Feasibility of adaptive sampling demonstrated at OHMX.bio
Recently, Oxford Nanopore Technologies released a new generation of its direct RNA sequencing kit, together with the first dedicated flow cell for RNA. Together, this increased the sequencing output and read accuracy dramatically, but moreover, the translocation speed for RNA raised to around 130 bases per second. With these improvements included, OHMX.bio could for the first time demonstrate to reach an average enrichment factor of around 2.7 for a set of transcripts of interest in adaptive sampling for direct RNA. The read output of this approach enables a detailed study of the present isoforms and splicing patterns in these genes of interest (Figure 2). It also offers an enriched and direct readout of the RNA modifications and with the models trained for picking up these modifications ever improving, exciting times for epitranscriptomics are definitely on the horizon.
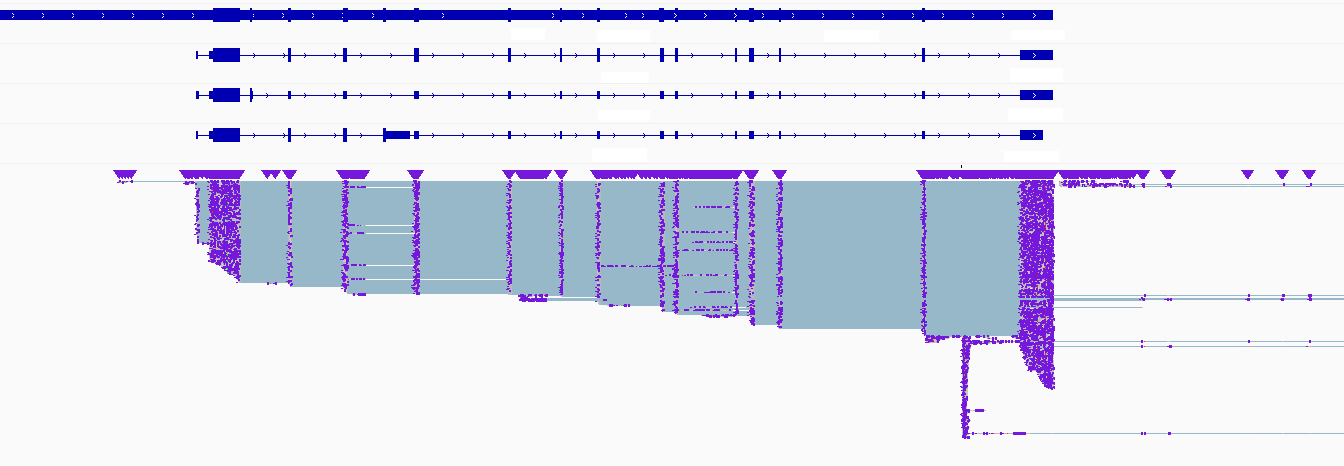
Figure 2: Genome browser view of a white-listed gene of interest. The applied adaptive sampling strategy allowed to enrich the sequencing of full-length native RNA molecules from a selection of genes of interest. The enhanced RNA coverage on this gene of interest boosts the analysis of splicing, isoforms and RNA modifications.
The OHMX.bio approach
OHMX.bio has a specialty lab focused on several sequencing solutions. This includes both short- and long-read sequencing with a goal to provide a one-stop solution for your research. OHMX.bio can combine genomic and epigenomic data, sequenced in-house, with (epi)transcriptomic and even translatomic and proteomic data. This means OHMX.bio can provide complete information about the pathway of a gene, from its exact genetic code (and mutations) to its regulation, expression, and translation.
The advantage of using one lab to perform all analyses, in a pick-and-mix fashion, is the guarantee that all samples are processed within the same quality system, by the same experts. This leads to fewer logistic errors and paperwork and allows for clear answers. Whether it is for oncological research or biomarker discovery, OHMX.bio can help.
Do you have questions about adaptive sampling?
Fill out the form below and our experts will get back to you as soon as possible!
References
[1] A. Payne, N. Holmes, T. Clarke, R. Munro, B. J. Debebe, and M. Loose, “Readfish enables targeted nanopore sequencing of gigabase-sized genomes,” Nat Biotechnol, 2020, doi: 10.1038/s41587-020-00746-x.
[2] L. Weilguny et al., “Dynamic, adaptive sampling during nanopore sequencing using Bayesian experimental design,” Nat Biotechnol, vol. 41, no. 7, pp. 1018–1025, Jul. 2023, doi: 10.1038/s41587-022-01580-z.
[3] S. Martin, D. Heavens, Y. Lan, S. Horsfield, M. D. Clark, and R. M. Leggett, “Nanopore adaptive sampling: a tool for enrichment of low abundance species in metagenomic samples,” Genome Biol, vol. 23, no. 1, Dec. 2022, doi: 10.1186/s13059-021-02582-x.
[4] T. R. Gingeras, “Current frontiers in RNA research,” Frontiers in RNA Research, vol. 1, May 2023, doi: 10.3389/frnar.2023.1152146.
[5] J. Cerneckis, G. L. Ming, H. Song, C. He, and Y. Shi, “The rise of epitranscriptomics: recent developments and future directions,” Trends in Pharmacological Sciences, vol. 45, no. 1. Elsevier Ltd, pp. 24–38, Jan. 01, 2024. doi: 10.1016/j.tips.2023.11.002.
[6] I. S. Naarman-De Vries, E. Gjerga, C. L. A. Gandor, and C. Dieterich, “Adaptive Sampling as tool for Nanopore direct RNA-sequencing,” BioRxiv, 2022, doi: 10.1101/2022.10.14.512223.
